About PhytoLoss
General objectives
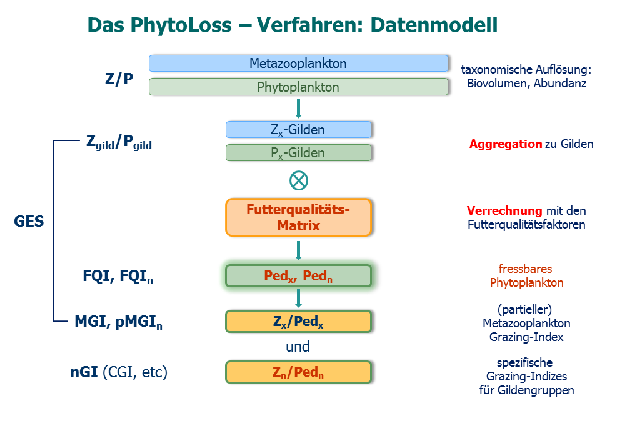
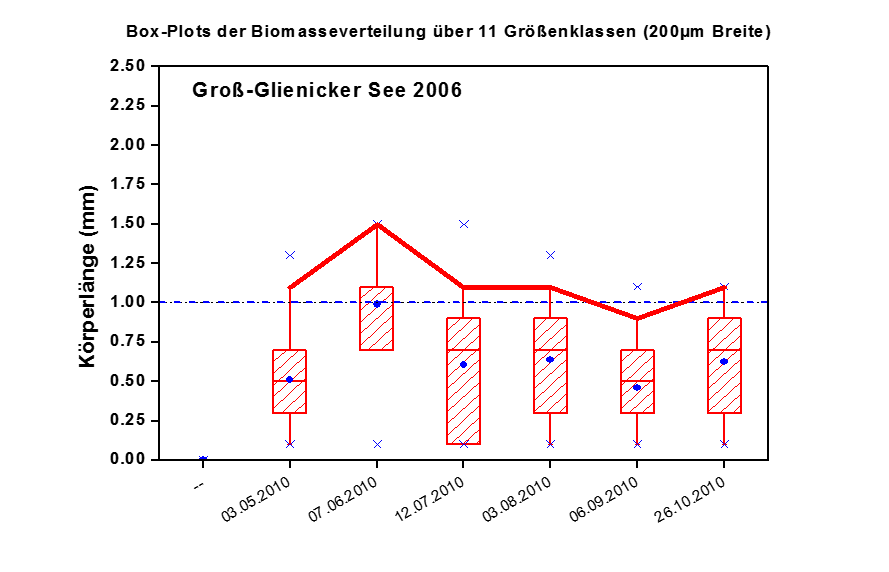
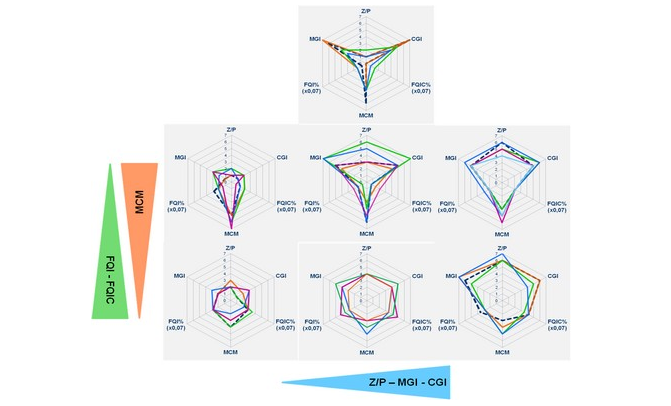
PhytoLoss is a database modul for the assessment of ecological quality in fresh waters using zooplankton data. PhytoLoss estimates the influence of food web interactions on phytoplankton biomass and trophic state. Funding is provided by LAWA (LänderArbeits-gemeinschaft WAsser).

Why PhytoLoss?
Platform for zooplankton analyses
PhytoLoss aims at the compensation of negative effects on zooplankton analysis from of the exclusion of the EU Water Framework Directive. It provides tools for data import, linking with phytoplankton data and calculation of index values.
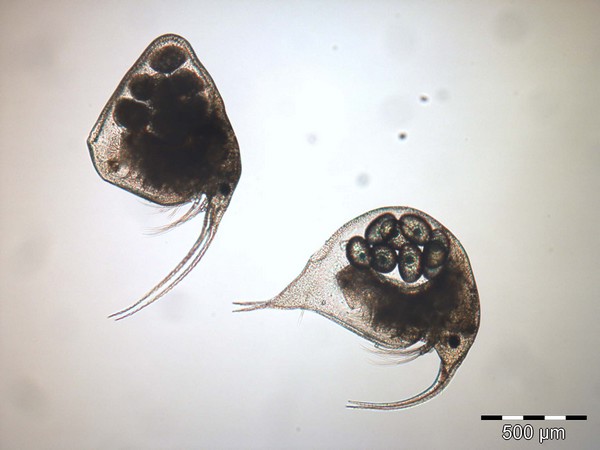
OTL-MZ
Operational Taxa List Metazooplankton
The OTL-MZ is necessary for consistent coding of zooplankton taxa according to international taxonomic standards prior to the data import procedure. Thus, provided data remain valid for future analyses.
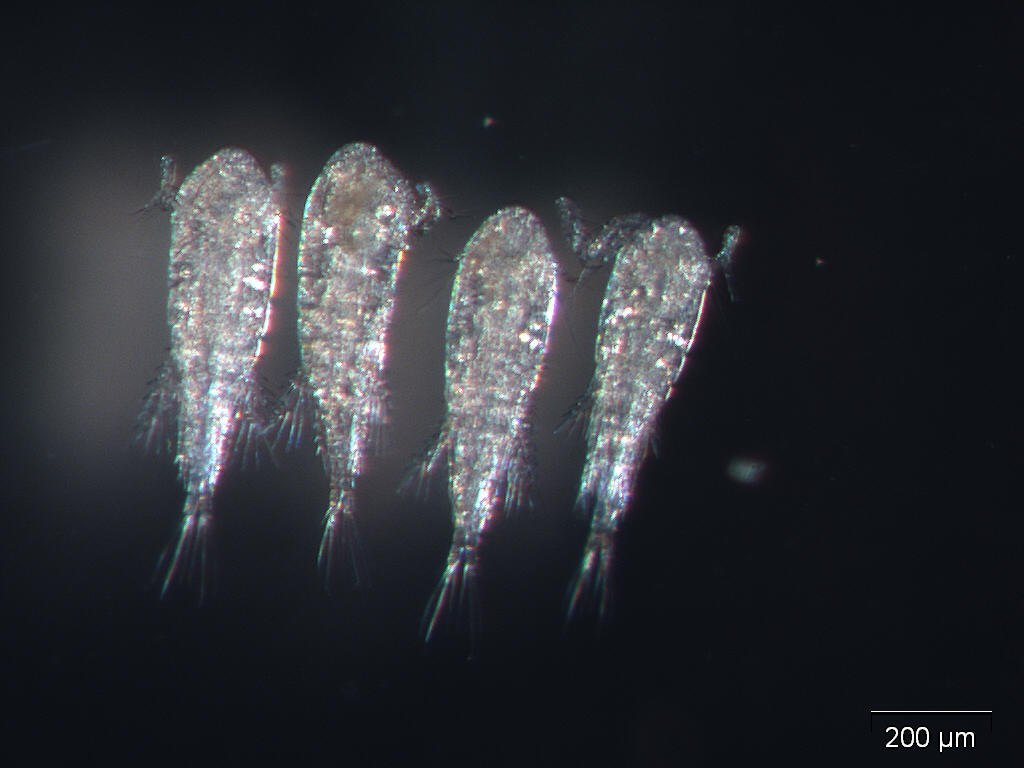
Data quality matters
Standardized import forms
Zooplankton analyses are essential for long-term monitoring of fish as well as the impact of climate change. Therefore, data quality management has to ensure future usability by implementing standard procedures and forms.